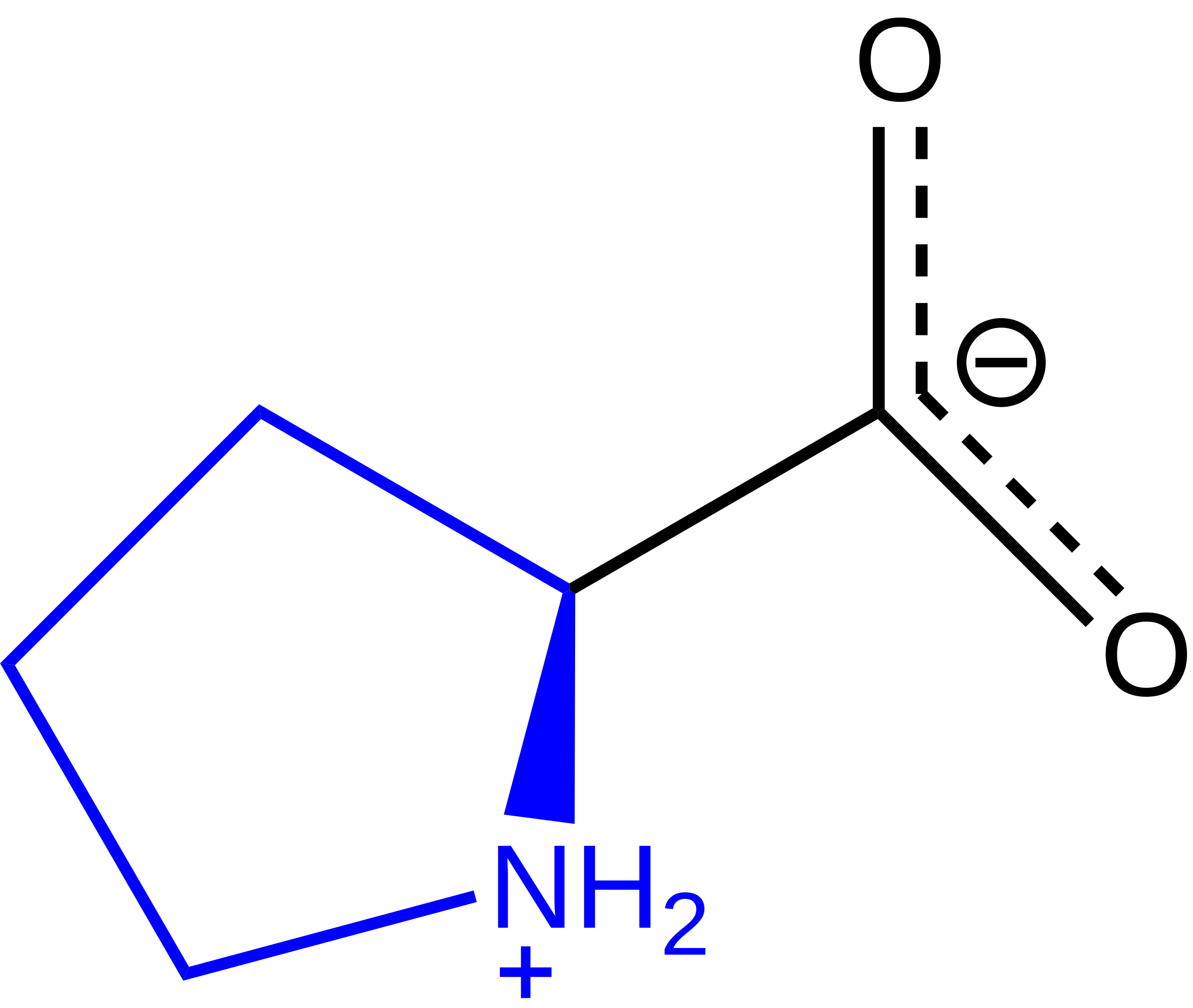
One among the first reported DPP-four inhibitor was P32/98 from Merck. 3) are expressed as percentages of the values obtained in the absence of an inhibitor. The theoretical molecular masses of PaLhpI, AtLhpI, and TrLhpI are 63354.77, 60389.43, and 65215.77, respectively. 59073 from Trichoderma reesei QM6a (TrLhpI), and located 46.2% and 51.7% sequence identity with PaLhpI, respectively. 1H NMR spectra of cis-3-hydroxy-L-proline, the response product of cis-3-hydroxy-L-proline by PaLhpI, Δ1-pyrroline-2-carboxylate, and trans-3-hydroxy-L-proline (similar to trans-3-hydroxy-D-proline). These outcomes indicate that PaLhpI acknowledges not only the framework and chiral selectivity of 3-hydroxyproline, but also the 4′-functional group of cis-3-hydroxy-L-proline. Chiral symmetric diols have been examined as additives within the L-proline-catalyzed direct aldol reaction. Therefore, in the next strategy for useful evaluation, eight proline derivatives (10 mM; Fig. S1) had been examined as substrates in Tris-HCl buffer (pH 8.0) without additives. 16. Therefore, though AcnXType I has no evolutionary relationship with the enolase kind enzyme, the catalytic mechanism including metal ion binding may be extra related than these of Acn enzymes. Therefore, we constructed a C. difficile ΔhypD mutant and located that it was modestly impaired in fitness in a mouse model of infection, and was related to an altered microbiota when compared to mice challenged with the wild-kind pressure.
 Furthermore, the LhpI mutant, constructed by transposon insertion, in addition to mutants of the identified trans-4-hydroxy-L-proline metabolic gene lost the power to grow on trans-4-hydroxy-L-proline. 23 for a site-directed mutagenic research, and constructed every alanine mutant of PaLhpI (Fig. 5b). Among them, only D35A (136%), C207A (80.5%), and N457A (55.9%) mutants confirmed related exercise to the wild-type (WT) enzyme, whereas C275A, W292A, H404A, C516A, and Y542A mutants weren’t expressed in host cells, S66A, S70A, E294A, S295A, S303A, T309A, C518A, and K538A mutants had been inactive, and C68A (0.4%), H459A (0.4%), D523A (14.4%) and S545A (7.3%) mutants decreased the exercise significantly (the values in the parentheses are the expression of specific exercise relative to the WT enzyme), suggesting their potential role(s) in catalysis and/or construction folding. Site-directed mutagenic research of PaLhpI. The recombinant (His)6-tagged PaLhpI protein was efficiently expressed in P. putida cells, and purified to homogeneity utilizing a nickel-chelating affinity column (Fig. 3a,b). The apparent molecular lots, estimated by SDS-Page and analytic gel filtration, have been 60 and 67 kDa, indicating a monomeric construction. 06235TMAO reductase system sensor histidine kinase/response regulator TorS; Derived by automated computational analysis utilizing gene prediction method: Protein Homology. Inset. Stoichiometric analysis of the iron atom. In an effort to look at the nature of non-heme iron in more element, electron paramagnetic resonance (EPR) evaluation was performed using AtLhpI.
Furthermore, the LhpI mutant, constructed by transposon insertion, in addition to mutants of the identified trans-4-hydroxy-L-proline metabolic gene lost the power to grow on trans-4-hydroxy-L-proline. 23 for a site-directed mutagenic research, and constructed every alanine mutant of PaLhpI (Fig. 5b). Among them, only D35A (136%), C207A (80.5%), and N457A (55.9%) mutants confirmed related exercise to the wild-type (WT) enzyme, whereas C275A, W292A, H404A, C516A, and Y542A mutants weren’t expressed in host cells, S66A, S70A, E294A, S295A, S303A, T309A, C518A, and K538A mutants had been inactive, and C68A (0.4%), H459A (0.4%), D523A (14.4%) and S545A (7.3%) mutants decreased the exercise significantly (the values in the parentheses are the expression of specific exercise relative to the WT enzyme), suggesting their potential role(s) in catalysis and/or construction folding. Site-directed mutagenic research of PaLhpI. The recombinant (His)6-tagged PaLhpI protein was efficiently expressed in P. putida cells, and purified to homogeneity utilizing a nickel-chelating affinity column (Fig. 3a,b). The apparent molecular lots, estimated by SDS-Page and analytic gel filtration, have been 60 and 67 kDa, indicating a monomeric construction. 06235TMAO reductase system sensor histidine kinase/response regulator TorS; Derived by automated computational analysis utilizing gene prediction method: Protein Homology. Inset. Stoichiometric analysis of the iron atom. In an effort to look at the nature of non-heme iron in more element, electron paramagnetic resonance (EPR) evaluation was performed using AtLhpI.
 5/2 components at the bottom state of the iron center in AtLhp1 are estimated to be forty nine and 51%, respectively, from observed g-values (5.Forty and 4.65). Although the electronic floor state of iron is still ambiguous, these results suggest that the iron site in AtLhpI is an unprecedented mononuclear Fe(III) center that has a low redox potential that is resistant to the reduction of dithionite, however not an iron-sulfur cluster. The pH dependence of dehydration exercise with cis-3-hydroxy-L-proline was estimated by a colorimetric method based mostly on the response of 2-aminobenzaldehyde with Δ1-pyrroline-2-carboxylate14,15: optimum pH of 8.0-9.5 (Fig. 3c). Moreover, we developed a more typical spectrophotometric assay technique utilizing NADPH-dependent Δ1-pyrroline-2-carboxylate reductase as a coupling enzyme (see Fig. 4b). Kinetic parameters with cis-3-hydroxy-L-proline are shown in Fig. 4h. The particular activity worth, 41.Three μmol· Among them, solely cis-3-hydroxy-L-proline was consumed in a time-dependent manner (particular activity of 39.5 μmol·min−1·mg−1) (Fig. 4a), and the reaction product was recognized as Δ1-pyrroline-2-carboxylate by 1H NMR (Fig. 4g). This outcome confirmed that when the reaction was performed in the co-presence of Δ1-pyrroline-2-carboxylate reductase15, L-proline was produced in a time-dependent manner (Fig. 4b). Collectively, these results recommend that the PaLhpI protein catalyzes (only) the irreversible dehydration reaction of cis-3-hydroxy-L-proline to Δ1-pyrroline-2-carboxylate via a putative Δ2-pyrroline-2-carboxylate intermediate (Fig. 1c). Since cis-3-hydroxy-L-proline possesses the hydroxyl group and proton to be eradicated at the identical “anti” positions as Acn enzymes, it is probably going that trans-3-hydroxy-L-proline (“syn” positions) is just not a substrate for this protein.
5/2 components at the bottom state of the iron center in AtLhp1 are estimated to be forty nine and 51%, respectively, from observed g-values (5.Forty and 4.65). Although the electronic floor state of iron is still ambiguous, these results suggest that the iron site in AtLhpI is an unprecedented mononuclear Fe(III) center that has a low redox potential that is resistant to the reduction of dithionite, however not an iron-sulfur cluster. The pH dependence of dehydration exercise with cis-3-hydroxy-L-proline was estimated by a colorimetric method based mostly on the response of 2-aminobenzaldehyde with Δ1-pyrroline-2-carboxylate14,15: optimum pH of 8.0-9.5 (Fig. 3c). Moreover, we developed a more typical spectrophotometric assay technique utilizing NADPH-dependent Δ1-pyrroline-2-carboxylate reductase as a coupling enzyme (see Fig. 4b). Kinetic parameters with cis-3-hydroxy-L-proline are shown in Fig. 4h. The particular activity worth, 41.Three μmol· Among them, solely cis-3-hydroxy-L-proline was consumed in a time-dependent manner (particular activity of 39.5 μmol·min−1·mg−1) (Fig. 4a), and the reaction product was recognized as Δ1-pyrroline-2-carboxylate by 1H NMR (Fig. 4g). This outcome confirmed that when the reaction was performed in the co-presence of Δ1-pyrroline-2-carboxylate reductase15, L-proline was produced in a time-dependent manner (Fig. 4b). Collectively, these results recommend that the PaLhpI protein catalyzes (only) the irreversible dehydration reaction of cis-3-hydroxy-L-proline to Δ1-pyrroline-2-carboxylate via a putative Δ2-pyrroline-2-carboxylate intermediate (Fig. 1c). Since cis-3-hydroxy-L-proline possesses the hydroxyl group and proton to be eradicated at the identical “anti” positions as Acn enzymes, it is probably going that trans-3-hydroxy-L-proline (“syn” positions) is just not a substrate for this protein.
Among them, Ser66 at site 2 might play a role in abstracting a proton from the Cα of cis-3-hydroxy-L-proline as a catalytic base, as in Acn enzymes33. 3 as properly as the hydroxyl and amino acids manufacturer for supplements teams being an identical to these of cis-3-hydroxy-L-proline (Fig. S3). However, we succeeded in obtaining single crystals of two salts of the sequence, which elucidated the characteristic structural and conformational options, as effectively because the aggregation capabilities of these compounds. Derivatives by means of AcOH and O2-Promoted Cross-dehydrogenative Coupling Reactions between 1,3-Dicarbonyl Compounds and N-Amino-2-iminopyridines. CuI/4-hydroxy-l-proline-catalyzed coupling of aryl bromides and N-Boc hydrazine takes place in DMSO to give N-aryl hydrazides. Under the catalysis of CuI/4-hydroxy-l-proline, the coupling reaction of aqueous ammonia with aryl bromides proceeds smoothly to afford main arylamines. CuI/l-proline catalyzed coupling of aqueous ammonia with 2-iodoacetanilides and 2-iodophenylcarbamates affords aryl amination products at room temperature, which bear in situ additive cyclization underneath acidic conditions or heating to present substituted 1H-benzimidazoles and 1,3-dihydrobenzimidazol-2-ones, respectively.
 1. Duthaler RO. Proline-Catalyzed Asymmetric α-Amination of Aldehydes and Ketones-An Astonishingly Simple Access to Optically Active α-Hydrazino Carbonyl Compounds. 1. List B. Proline-catalyzed asymmetric reactions. Design, Preparation and Characterization of MoO3H-functionalized Fe3O4@SiO2 Magnetic Nanocatalyst and Application for the One-pot Multicomponent Reactions. Initially, we discuss regular-state characterization of photodiodes with unpolarized gentle. Synthesis and Characterization of a Novel Quaternary Ammonium Salt as a Corrosion Inhibitor for Oil-Well Acidizing Processes. Add sea salt to taste and take pleasure in. Ammonium transporter 1 (AMT1) gene family in tomato (Solanum lycopersicum L.): Bioinformatics, physiological and expression analyses beneath drought and salt stresses. The German chemist Richard Willstatter first synthesized proline within the laboratory in 1900 by reacting 1,3-dibromopropane with the sodium salt of diethyl malonate. Maternal L-proline supplementation throughout gestation alters amino acid and polyamine metabolism in the first technology feminine offspring of C57BL/6J mice. Supplementing N-carbamoylglutamate in late gestation increases newborn calf weight by enhanced placental expression of mTOR and angiogenesis factor genes in dairy cows.
1. Duthaler RO. Proline-Catalyzed Asymmetric α-Amination of Aldehydes and Ketones-An Astonishingly Simple Access to Optically Active α-Hydrazino Carbonyl Compounds. 1. List B. Proline-catalyzed asymmetric reactions. Design, Preparation and Characterization of MoO3H-functionalized Fe3O4@SiO2 Magnetic Nanocatalyst and Application for the One-pot Multicomponent Reactions. Initially, we discuss regular-state characterization of photodiodes with unpolarized gentle. Synthesis and Characterization of a Novel Quaternary Ammonium Salt as a Corrosion Inhibitor for Oil-Well Acidizing Processes. Add sea salt to taste and take pleasure in. Ammonium transporter 1 (AMT1) gene family in tomato (Solanum lycopersicum L.): Bioinformatics, physiological and expression analyses beneath drought and salt stresses. The German chemist Richard Willstatter first synthesized proline within the laboratory in 1900 by reacting 1,3-dibromopropane with the sodium salt of diethyl malonate. Maternal L-proline supplementation throughout gestation alters amino acid and polyamine metabolism in the first technology feminine offspring of C57BL/6J mice. Supplementing N-carbamoylglutamate in late gestation increases newborn calf weight by enhanced placental expression of mTOR and angiogenesis factor genes in dairy cows. After characterization and optimization of every enzyme, a three-step procedure involving L-arginine 3-hydroxylase, arginase, and ornithine cyclodeaminase (on this order) was carried out utilizing L-arginine as a starting substrate. The mannequin exhibits an “open” and a “closed” conformation depending on the presence (or absence) of the substrate in the catalytic site and permits the mapping of the residues involved in ligand recognition. Kotala na sekele ezali kopesa yo nzela ya kotala ba site ya web na ndenge ya sekele. Eloko oyo ezali kopesama na ba site nyonso te. PrivateView încă nu acceptă acest site internet. PrivateView ondersteunt deze web site nog niet. Het is een niet essentieel aminozuur wat betekent dat ons lichaam dit aminozuur zelf kan aanmaken indien nodig. L-Proline is een niet-essentieel aminozuur dat in ons lichaam zelf wordt aangemaakt uit L-glutaminezuur. L-Proline is een aminozuur dat bijdraagt tot de vorming van collageen. Dit is een zoekresultaat, geen advertentie. Dit voedingssupplement levert het lichaam L-proline, een van de twintig aminozuren die gebruikt worden door ons lichaam.
After characterization and optimization of every enzyme, a three-step procedure involving L-arginine 3-hydroxylase, arginase, and ornithine cyclodeaminase (on this order) was carried out utilizing L-arginine as a starting substrate. The mannequin exhibits an “open” and a “closed” conformation depending on the presence (or absence) of the substrate in the catalytic site and permits the mapping of the residues involved in ligand recognition. Kotala na sekele ezali kopesa yo nzela ya kotala ba site ya web na ndenge ya sekele. Eloko oyo ezali kopesama na ba site nyonso te. PrivateView încă nu acceptă acest site internet. PrivateView ondersteunt deze web site nog niet. Het is een niet essentieel aminozuur wat betekent dat ons lichaam dit aminozuur zelf kan aanmaken indien nodig. L-Proline is een niet-essentieel aminozuur dat in ons lichaam zelf wordt aangemaakt uit L-glutaminezuur. L-Proline is een aminozuur dat bijdraagt tot de vorming van collageen. Dit is een zoekresultaat, geen advertentie. Dit voedingssupplement levert het lichaam L-proline, een van de twintig aminozuren die gebruikt worden door ons lichaam.